invertiaDB Help Page
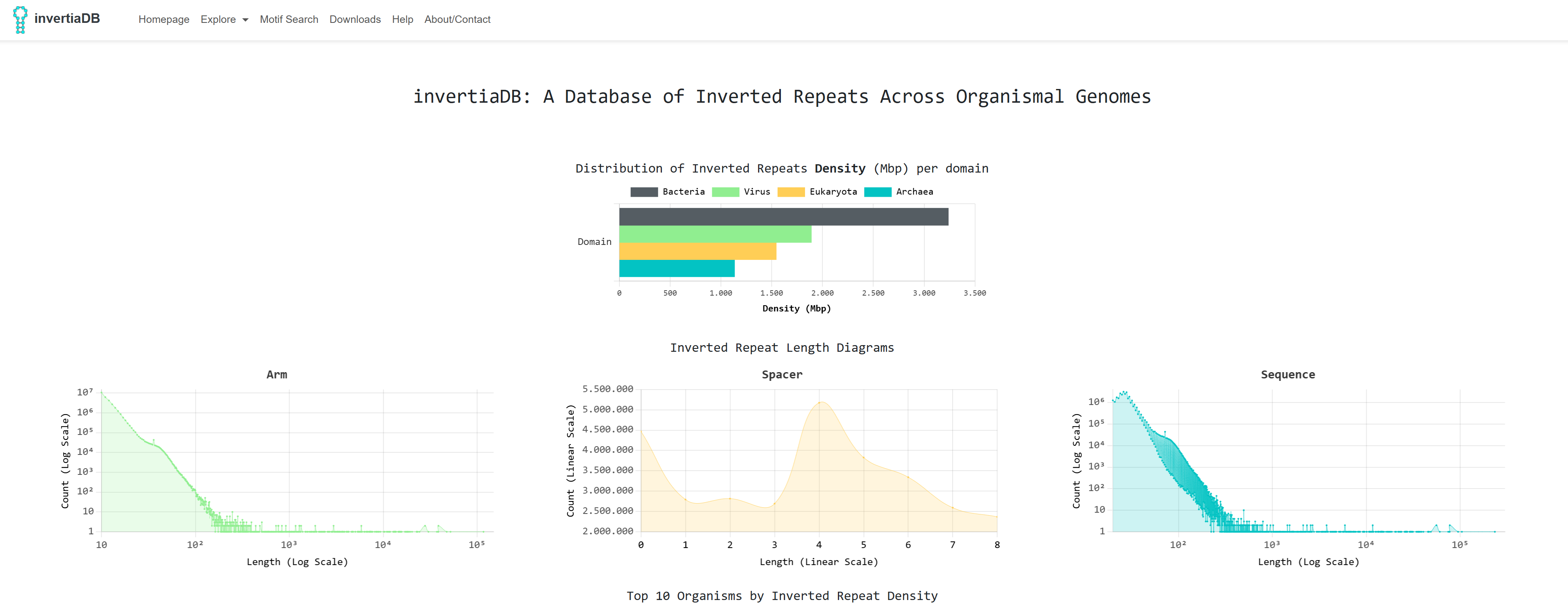
This is the homepage of invertiaDB. It is comprised of high level metadata about the
reasearch. The graphs of homepage are: A bar chart of the inverted repeats density per domain
, Three line chart that showcase the distribution of lengths for the arms, spacers and whole sequences
present on the dataset and
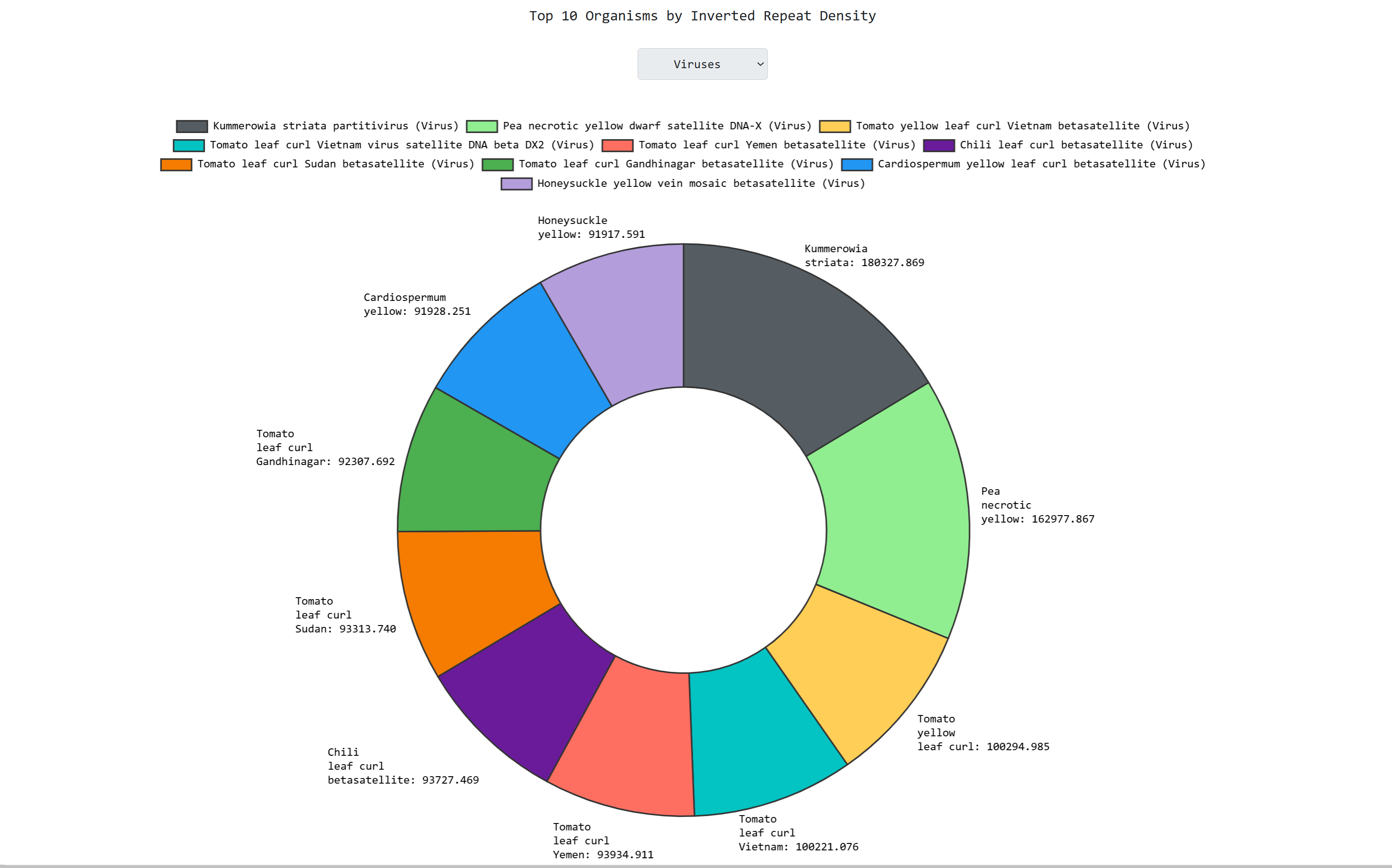
an interactive doughnut chart that graphs the top 10 most dense organisms in inverted repeats per
biological domain:
From here you can navigate to other pages of invertiaDB using the navigation bar.
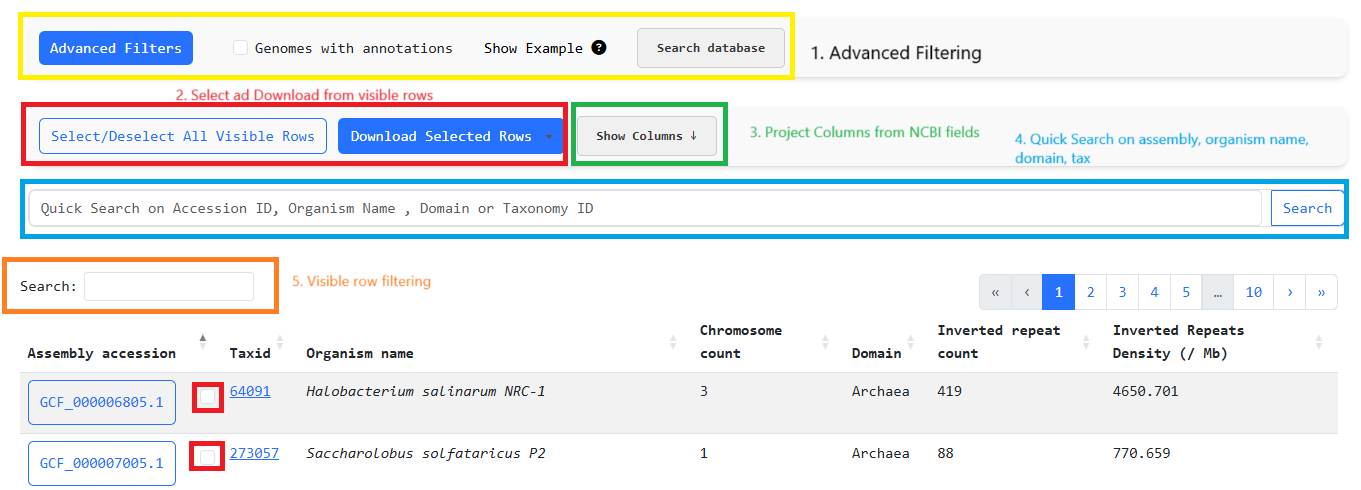
One of the core features of invertiaDB is the ability to search ( Explore -> Invertia Dataset ) via
various NCBI assembly genome fields as well
as inverted repeats metadata and filter out assemblies that you deem relevant for further analysis.
An example workflow would be to:
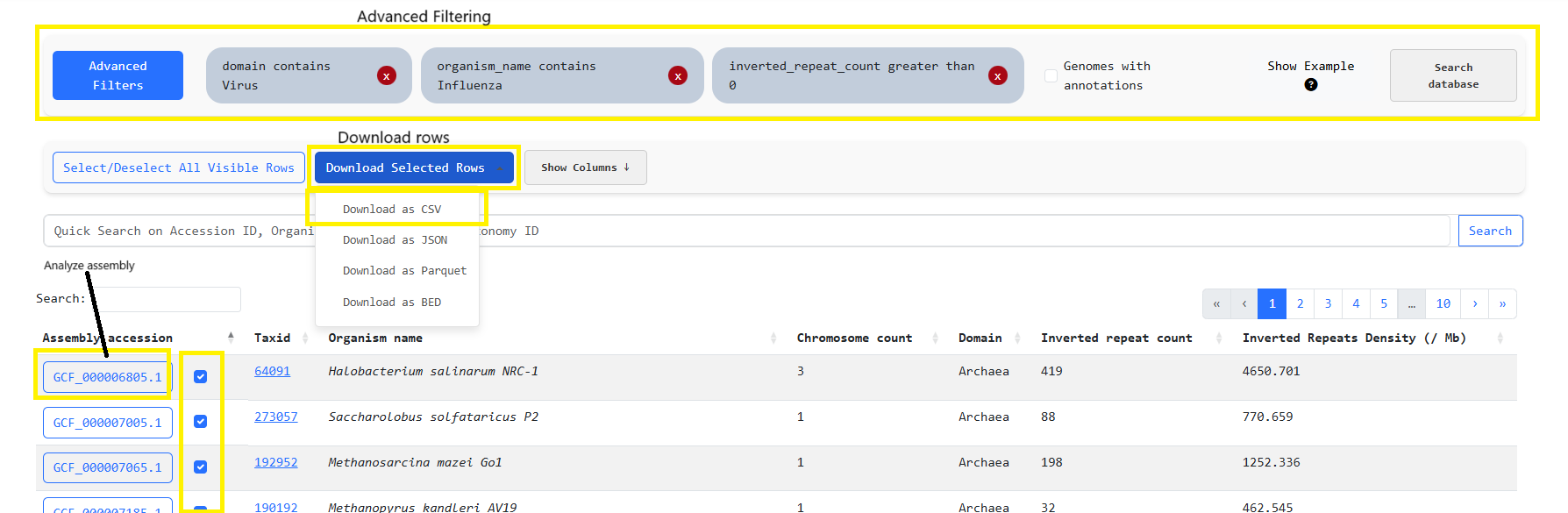
1. Quick search for the term "Influenza". This yields rows that come from the Bacterial and Viral
domain. So we must move on to advanced filtering
2. Perform advanced filtering to keep only the Viral rows through "Domain contains Virus" filter for
organisms that contain the word "Influenza" in their organism_name
3. Then we must keep only rows with IRs through the filter: "inverted_repeat_count greater than 0"
4. Finally we can select all rows and download the files for custom analysis or analyze the genomes
through invertia by clicking the blue button with their assembly_id.
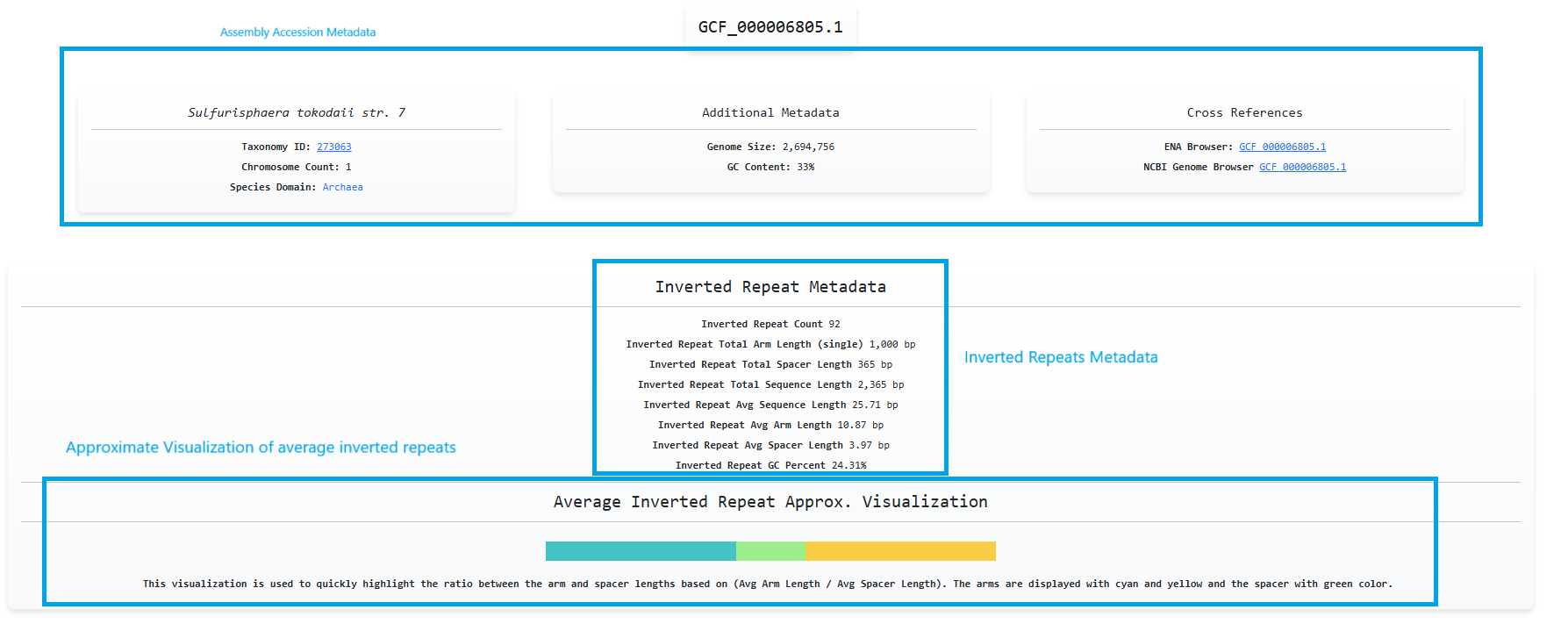
We can then click on the accession id button of a specific row and navigate to
the analysis page of this specific accession. Here we can see calculations on inverted repeats
and accession metadata with cross references to other public sources
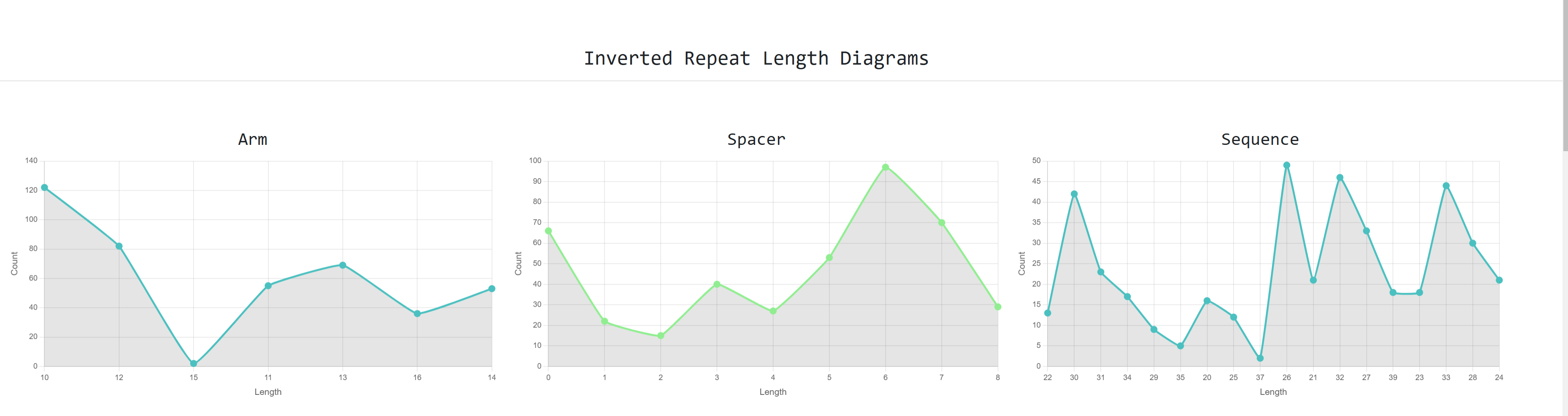
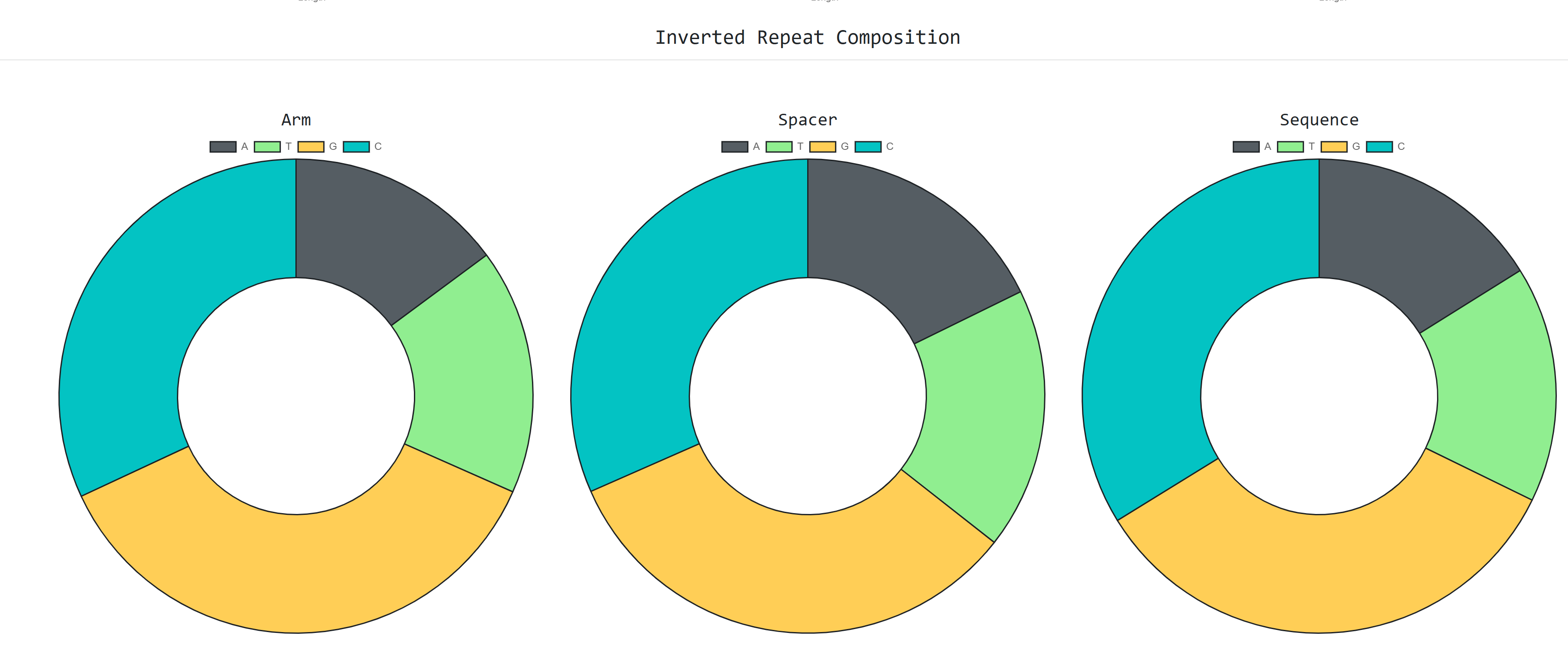
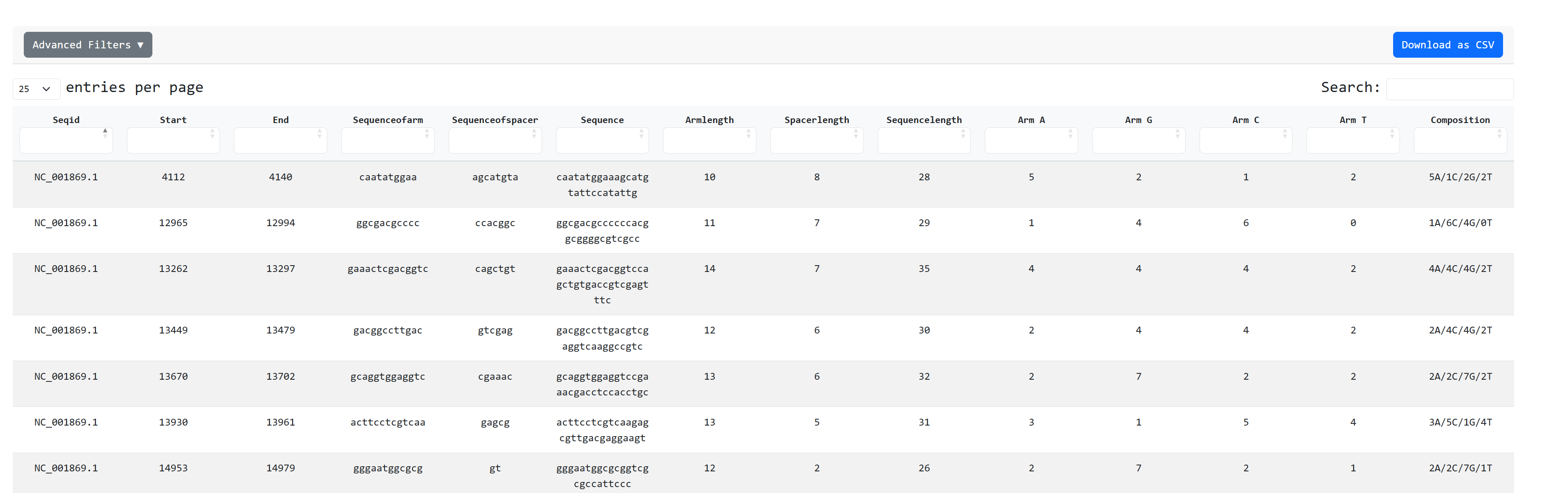
We can also see graphs about the distribution
on inverted repeat arm , spacer and sequence length and nucleotide composition charts
for the same sequences as well:
We can then inspect in tabular format the inverted repeats file as it is present on the dataset:
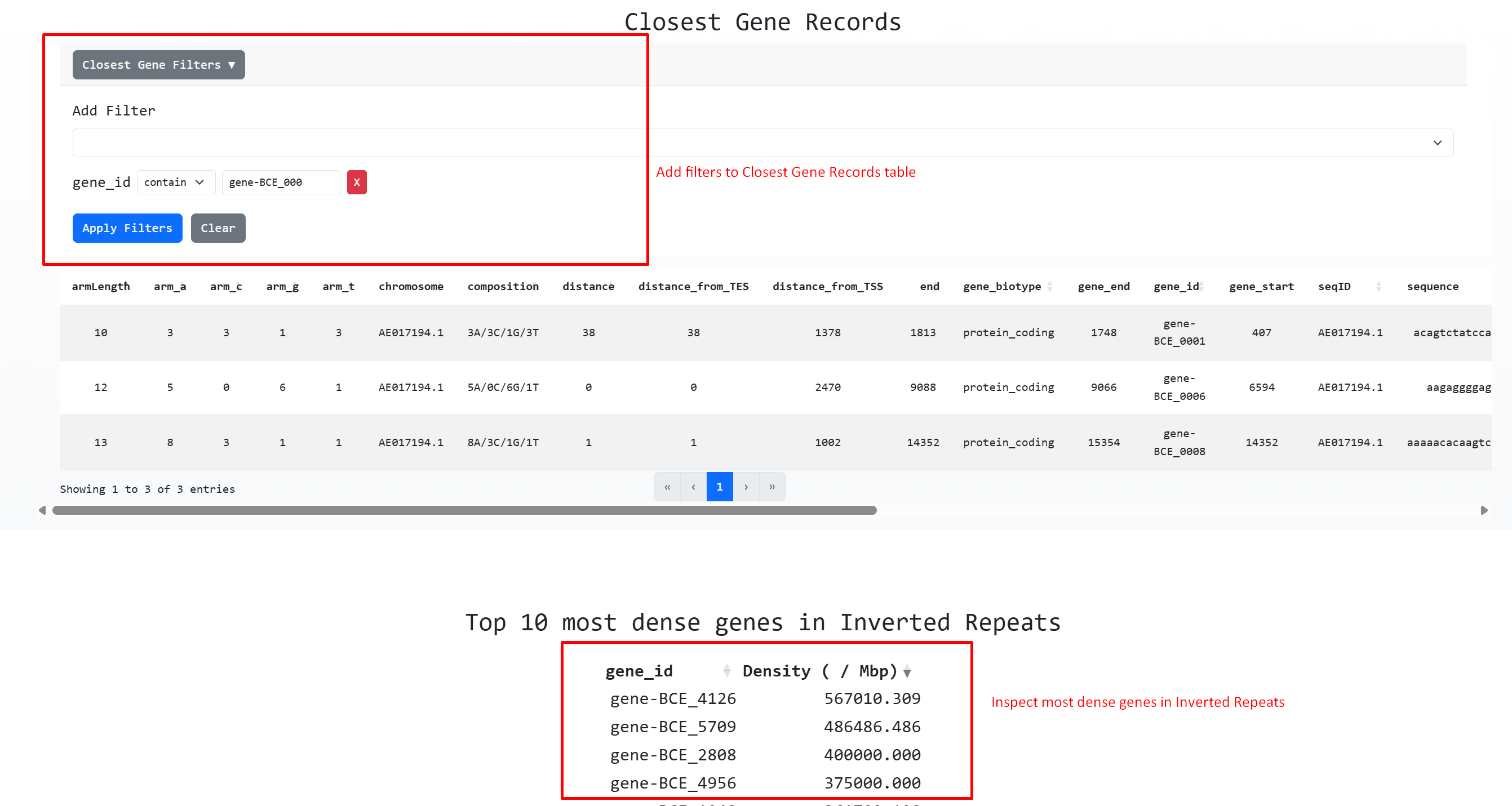
Finally if the genome was annotated through an NCBI gff file. We can use two gene related features
1. To search Inverted repeats that overlap with a gene based on its locus-tag ( Unique identifier )
2. To inspect the top 10 most dense genes in Inverted Repeats
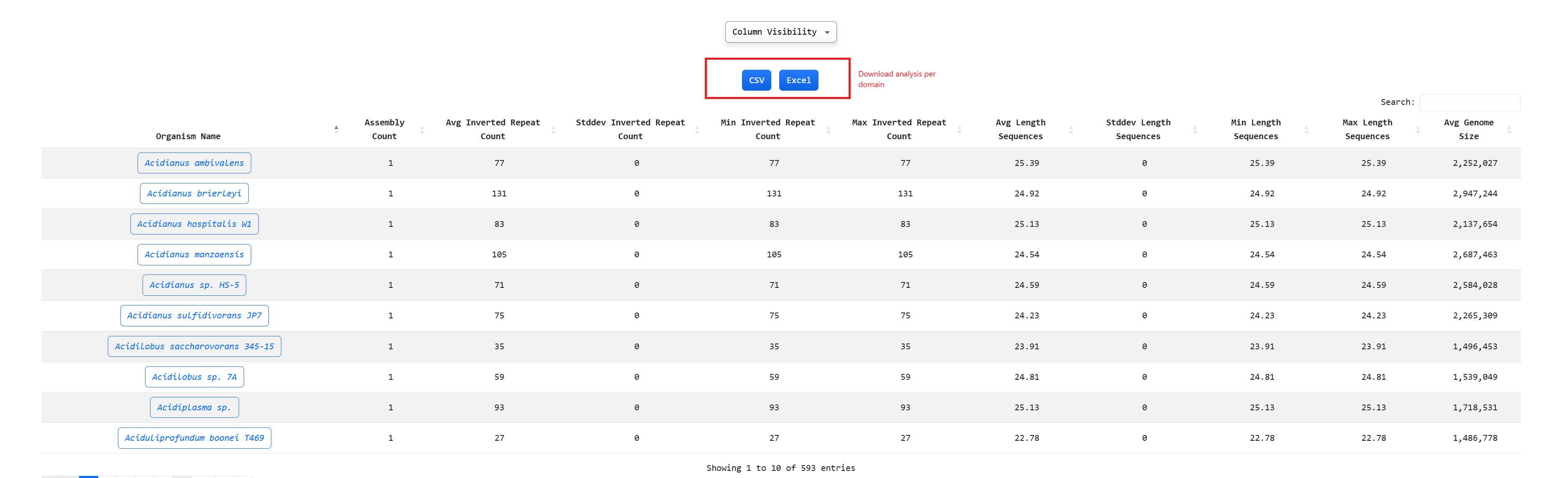
Through explore -> Domain we can perform an aggregated analysis per domain:
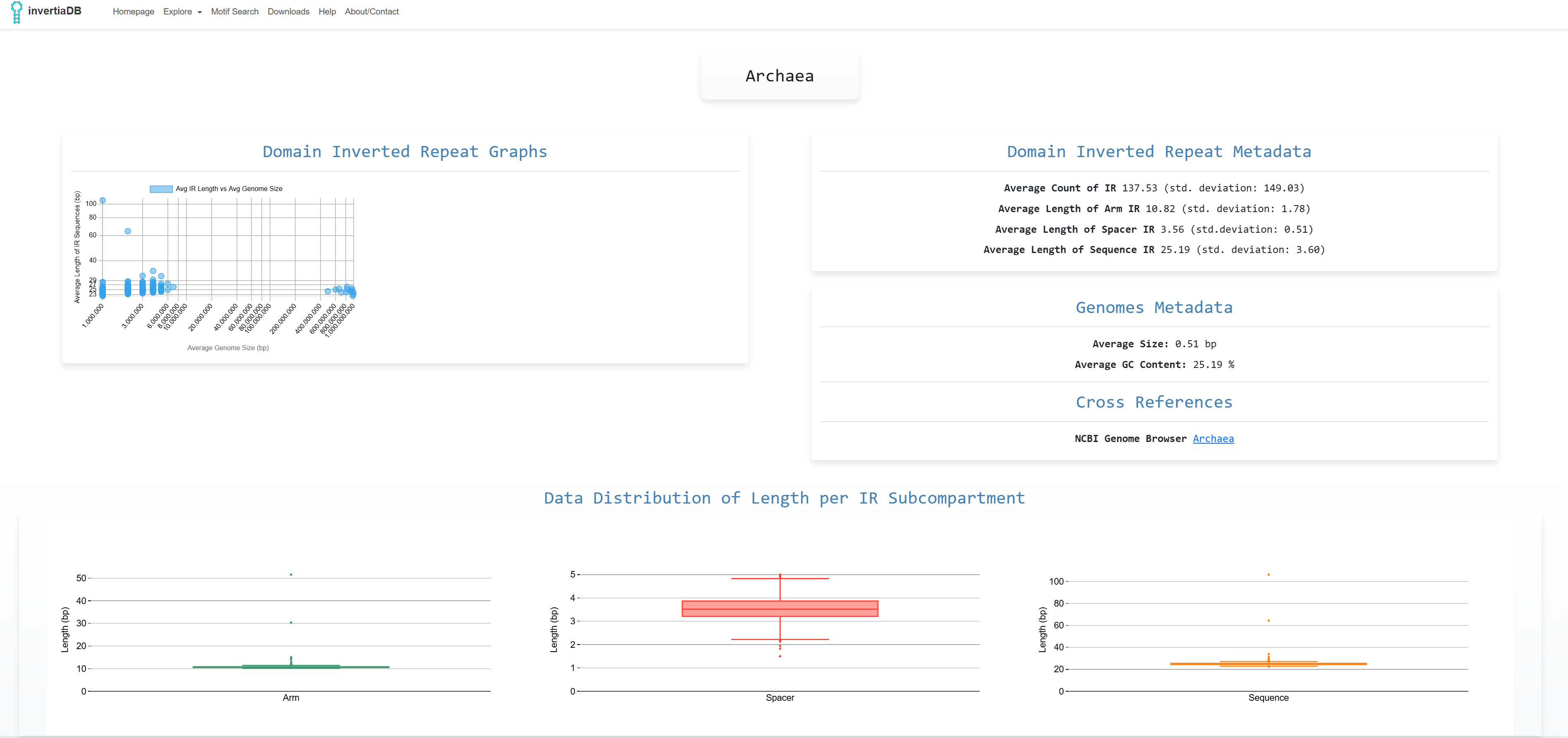
The aggregated analysis looks like these and there is the option to download the dataset in csv or xlsx
format. Also the graphs regarding Domain Inverted Repeat Graphs and Data Distributions of Length per IR
Subcompartment are presented:
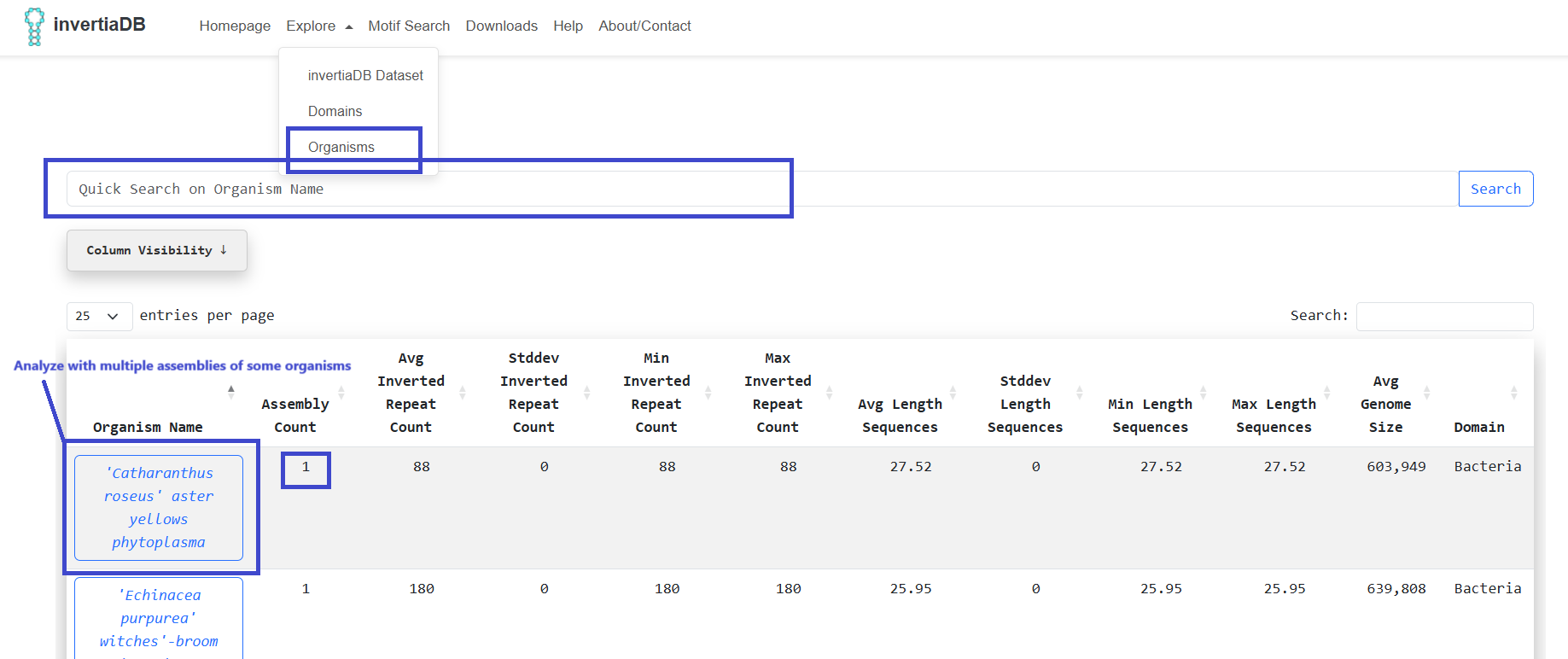
A more granular way to see the same statistics, meaning an aggregated analysis of multiple assemblies,
if the
an organism has more than 1, can be performed by the navbar option Explore -> Organisms:
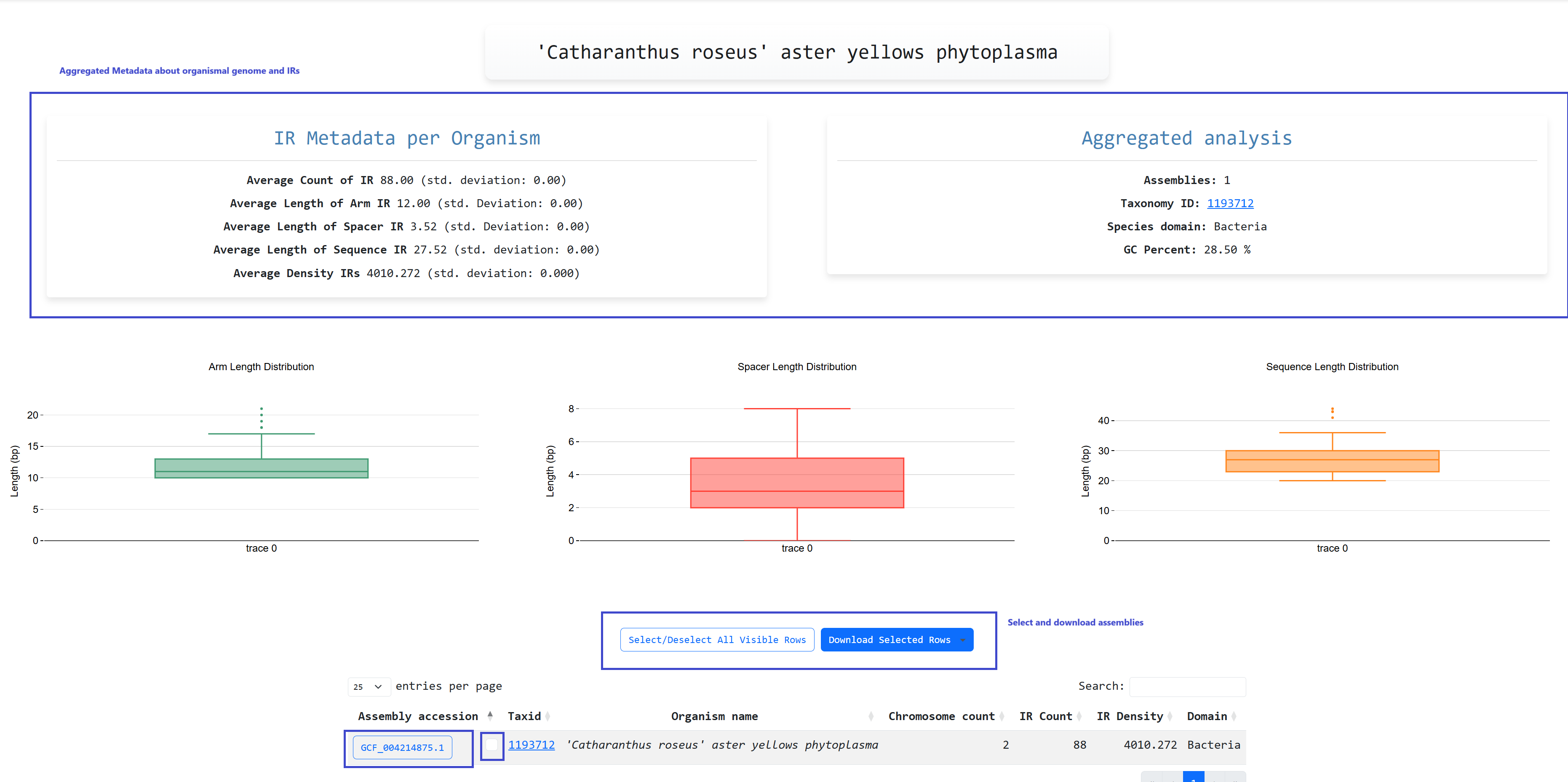
When clicking an organism that has multiple assemblies we are presented with an aggregated view of them
in
terms of
inverted repeats. We also have the ability to download and inspect specific assemblies the same as the
Invertia Dataset page. Similarly, Data Distributions of Length per IR
Subcompartment are presented.
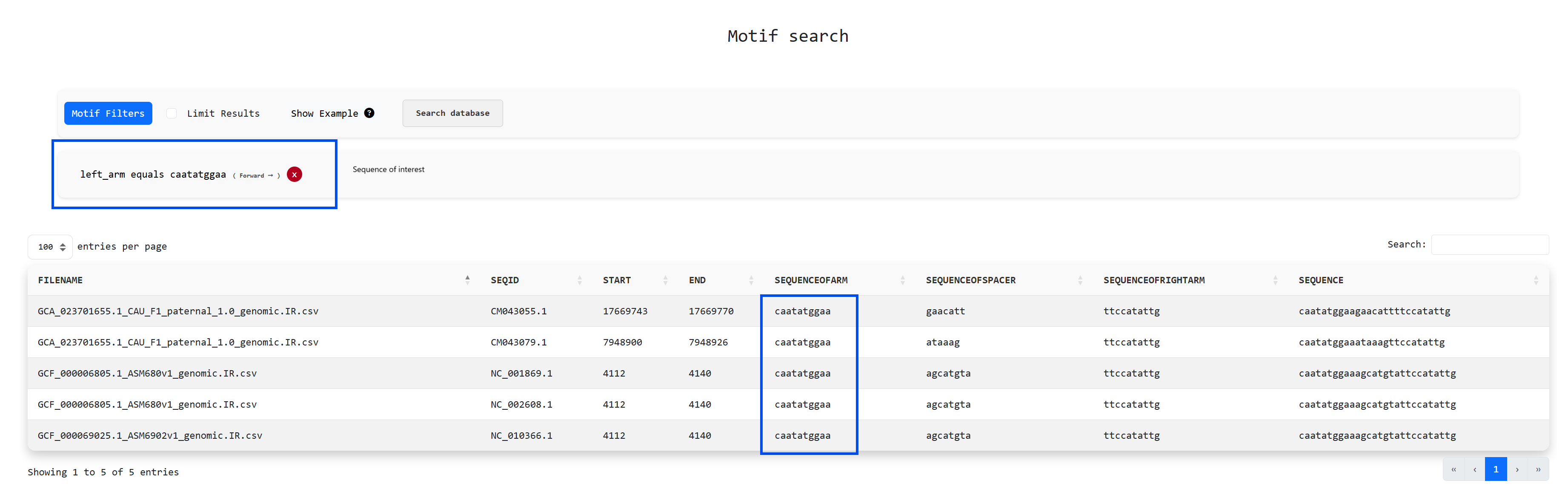
InvertiaDB also offers the ability to search for specific DNA motfis in the inverted repeat dataset
through the
navigation bar option
Motif Search. A trivial example use case that showcases this feature is to search for a sequence of
interest,
e.g. "caatatggaa" on the left arm of all inverted repeats:
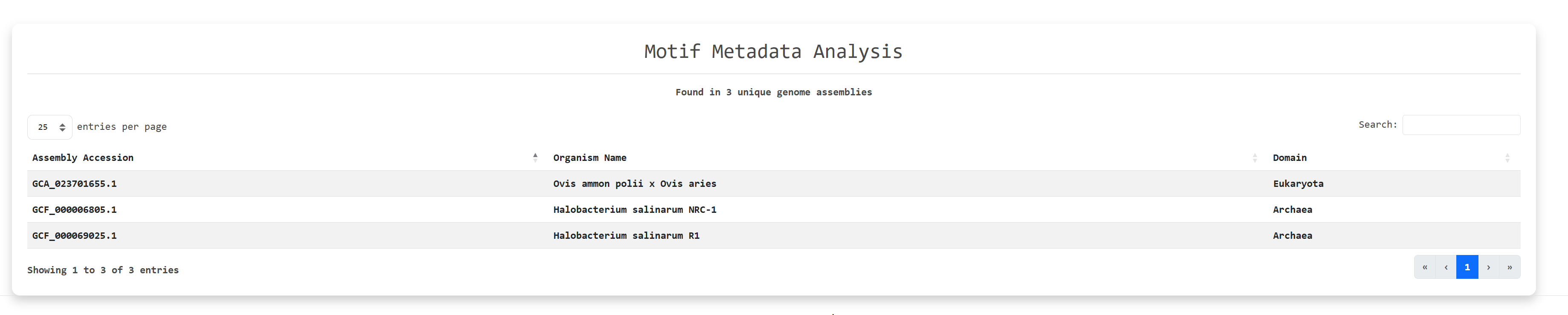
The primary search leads to a secondary search to find the files and the unique organisms in which these
motifs
were found to perform further analysis:
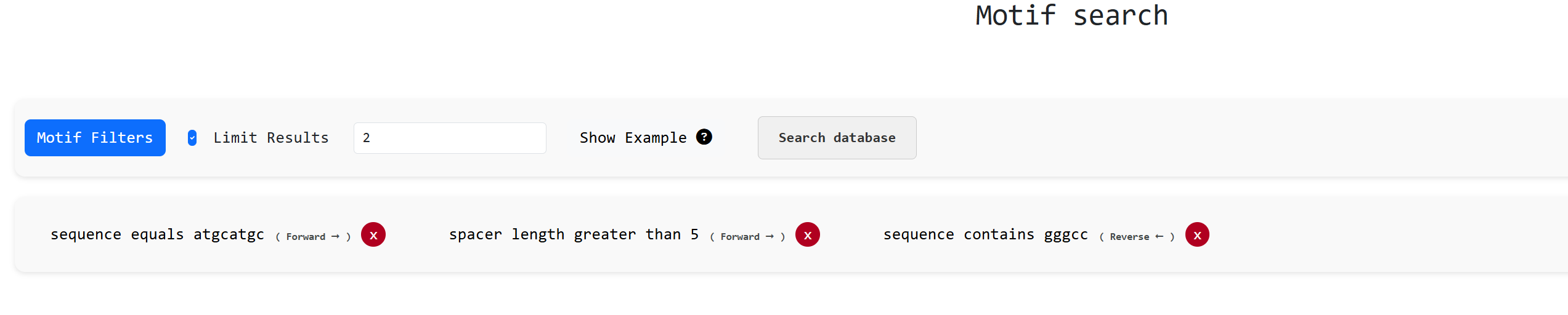
Apart from these use cases the motif search can be ran on various more advanced ways such as searching
both
arms , searching the reverse strand , both strands as well as apply text search functionalities
such as sequence contains X , starts with X and length comparisons such as "length equals X" or "length
greater than X" in complex ways while also having the ability to limit results. An example would look
like:
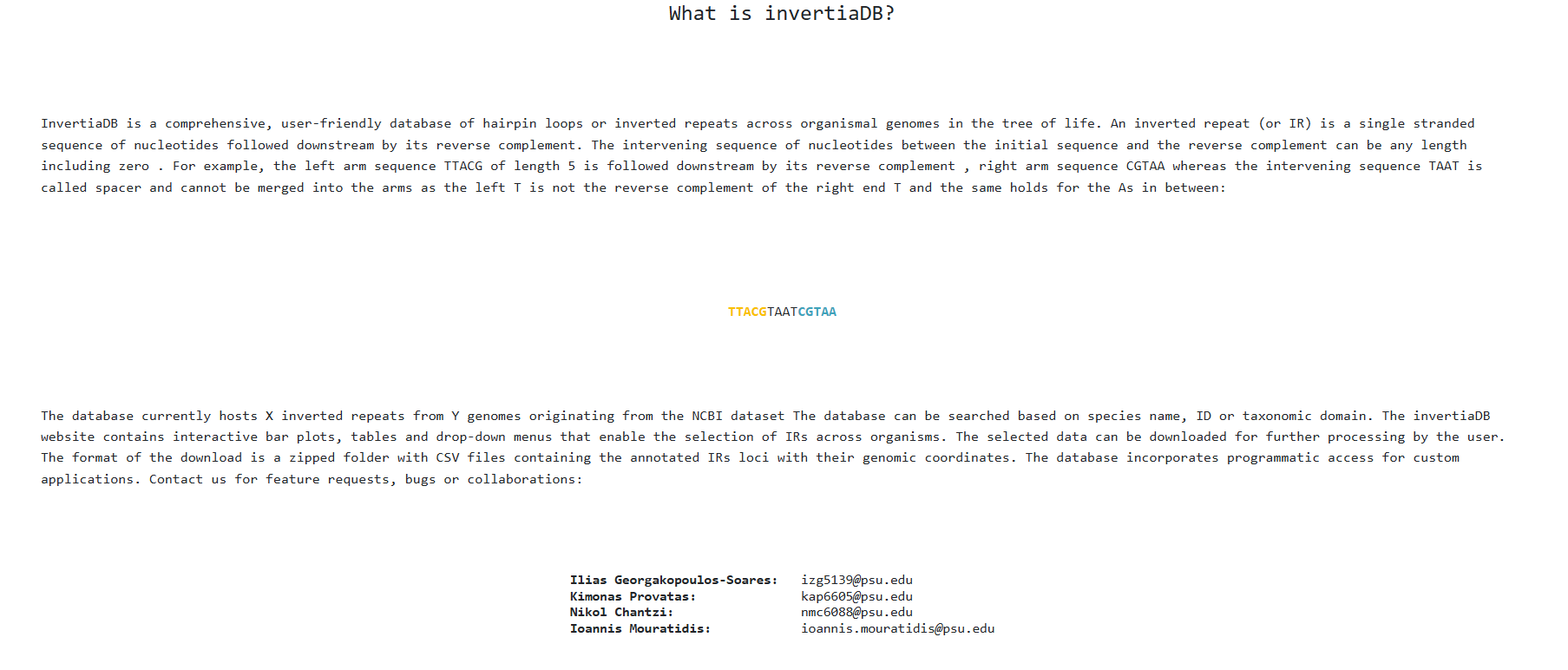
There is also an about/contact page that explains the basics of Inverted Repeats along with the lab's
researchers emails to
contact if you have any feedback or questions.
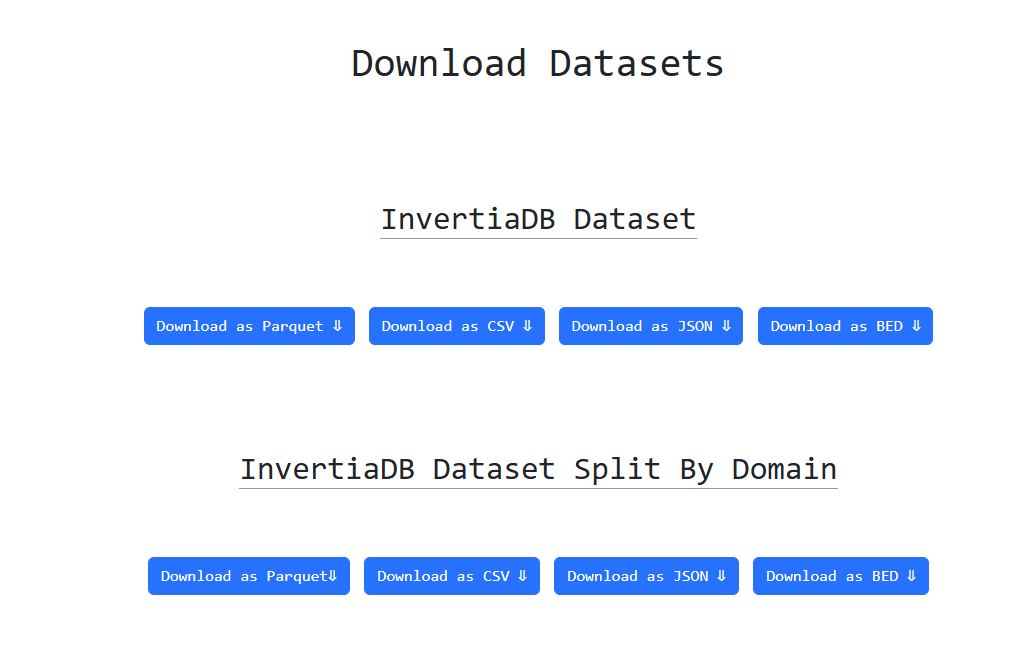
By navigating to Downloads through the navbar you have the option to download the whole dataset as a
whole or divided into
the four domains by clicking the respective button. There are four options in terms of data formats
namely csv, json, parquet and bed.